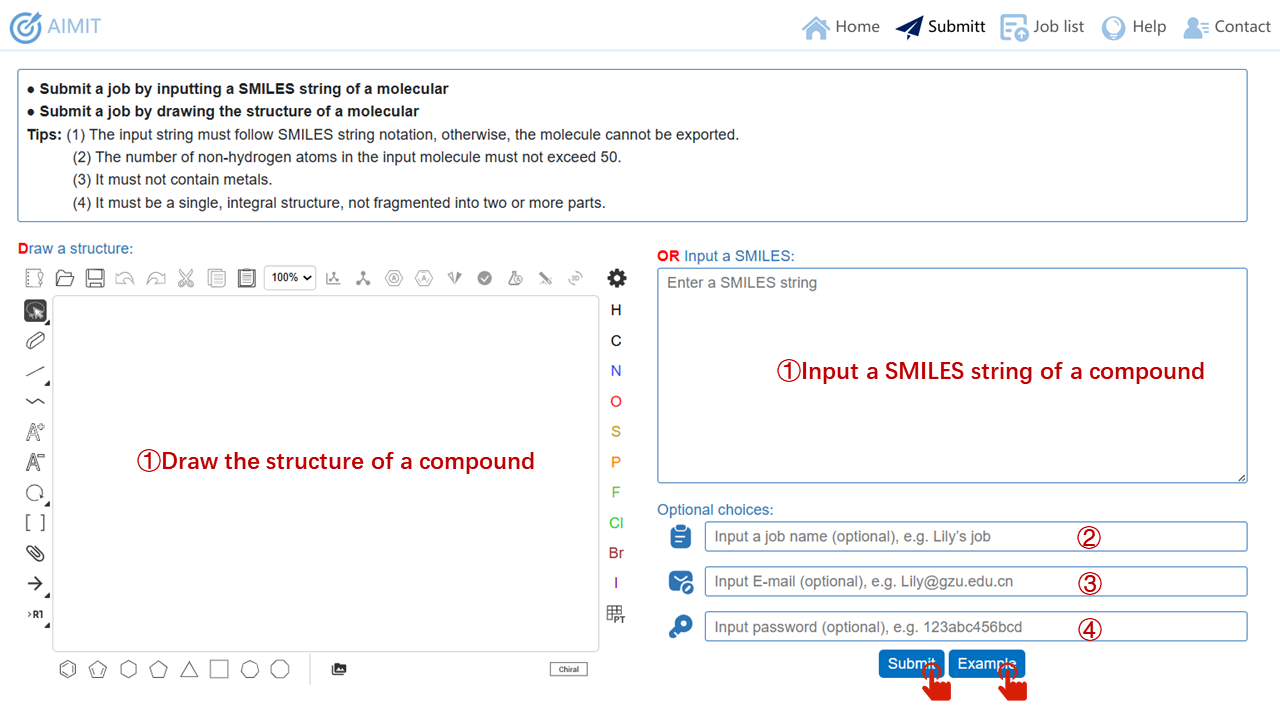
In Home webpage, users can click the Quick Start button or the Submit item in the navigation bar to jump to the Submit webpage. (1) AIMIT only requires users to input a SMILES string of a molecule or draw the structure of a molecule in the provided drawing interface to submit a task. (2) The input SMILES string must follow SMILES string notation, otherwise, the molecule cannot be exported. The number of non-hydrogen atoms in the input molecule must not exceed 50. It must not contain metals. It must be a single, integral structure, not fragmented into two or more parts. (3) Users also can name their jobs by their preferences and habits (This step is optional). (4) Users also can enter their email address and password (This step is optional). (5) When users click Submit button, job information will be submitted to back-end server. AIMIT will start computing as soon as possible. (6) If users need to check the example, users can click Example button to fill in example information, then launch the calculation.
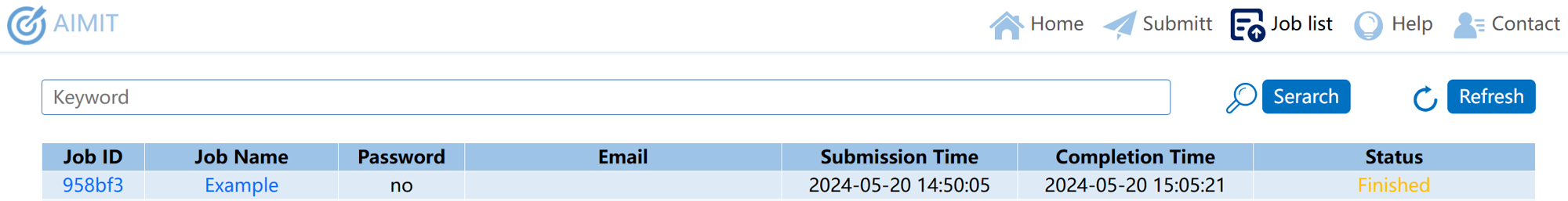
Once you submit the task, the webpage will redirect you to the job list page. After the task is completed, you can view the task results by clicking on the job ID, and job name.
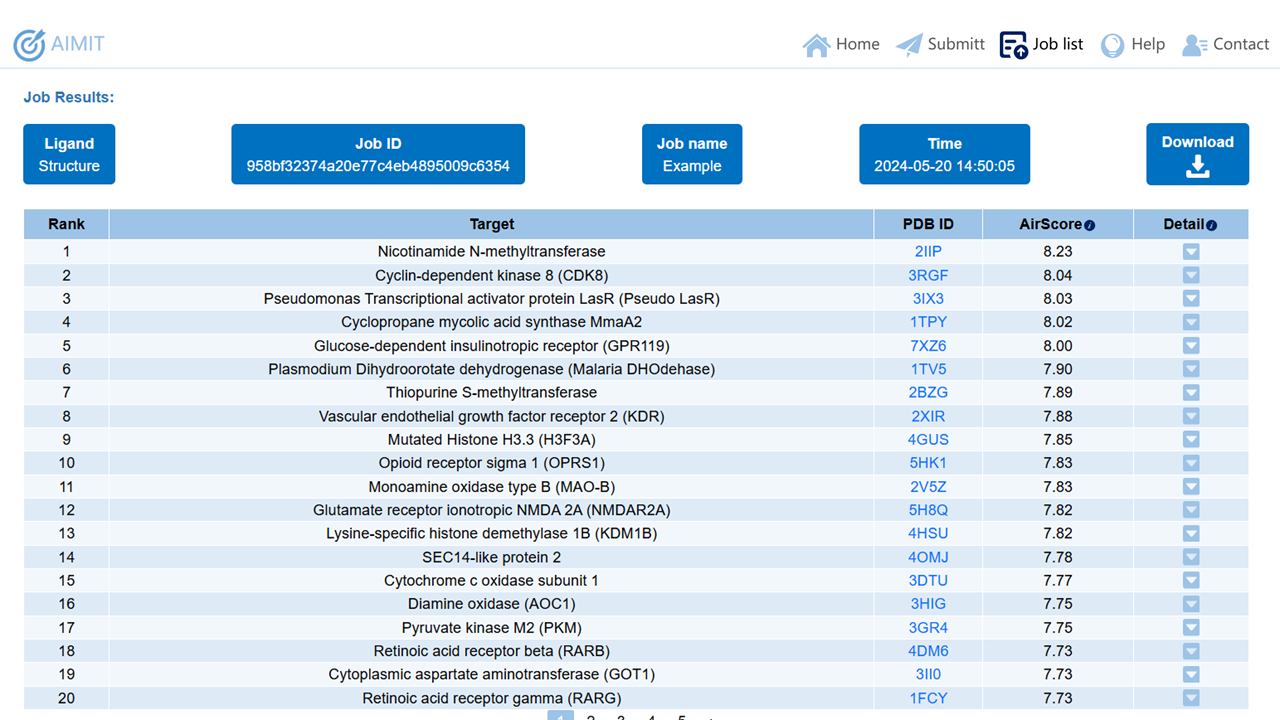
Once the task computation is completed, you will first receive a table displaying information about the targets, sorted from the highest to the lowest score. The specific information presented in the table includes the target name, the PDB ID of the target, and the specific scoring value.
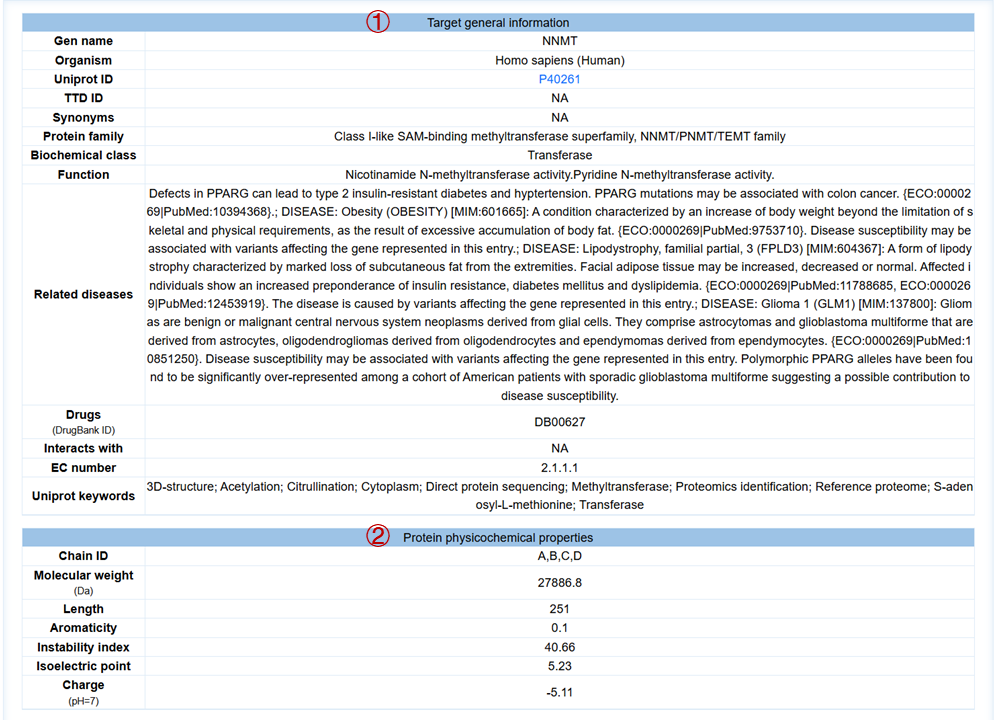
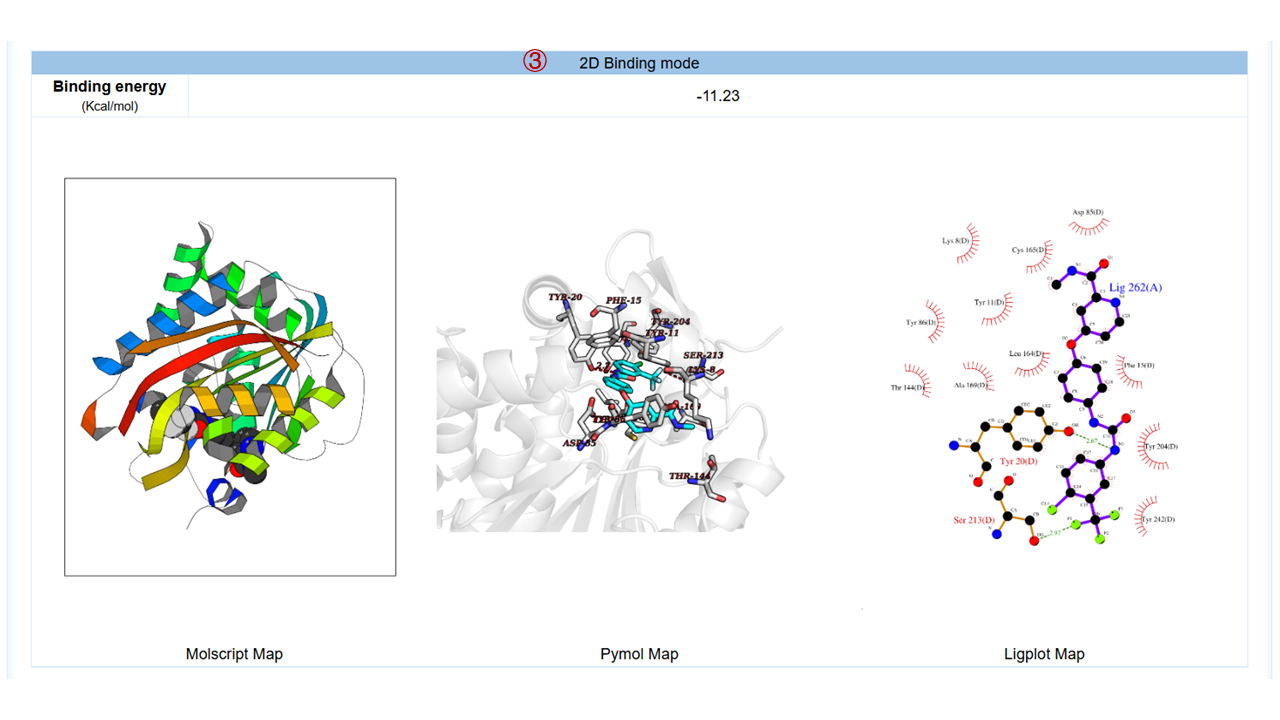
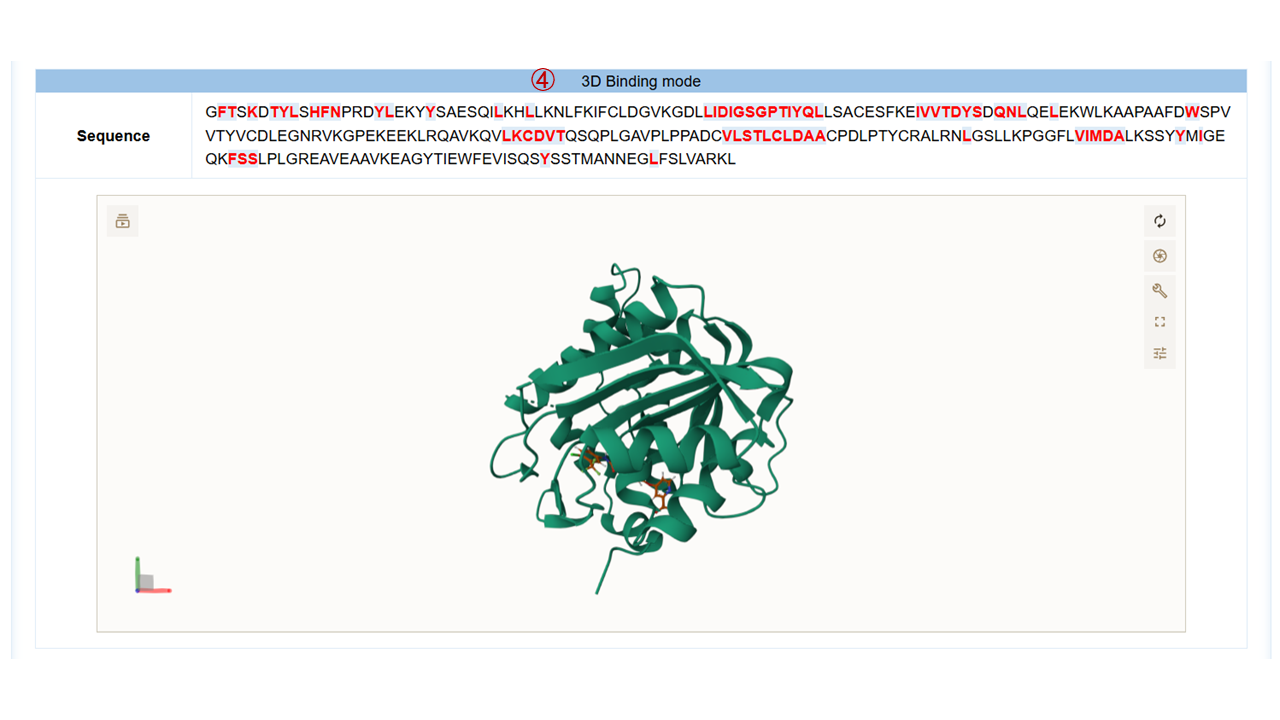
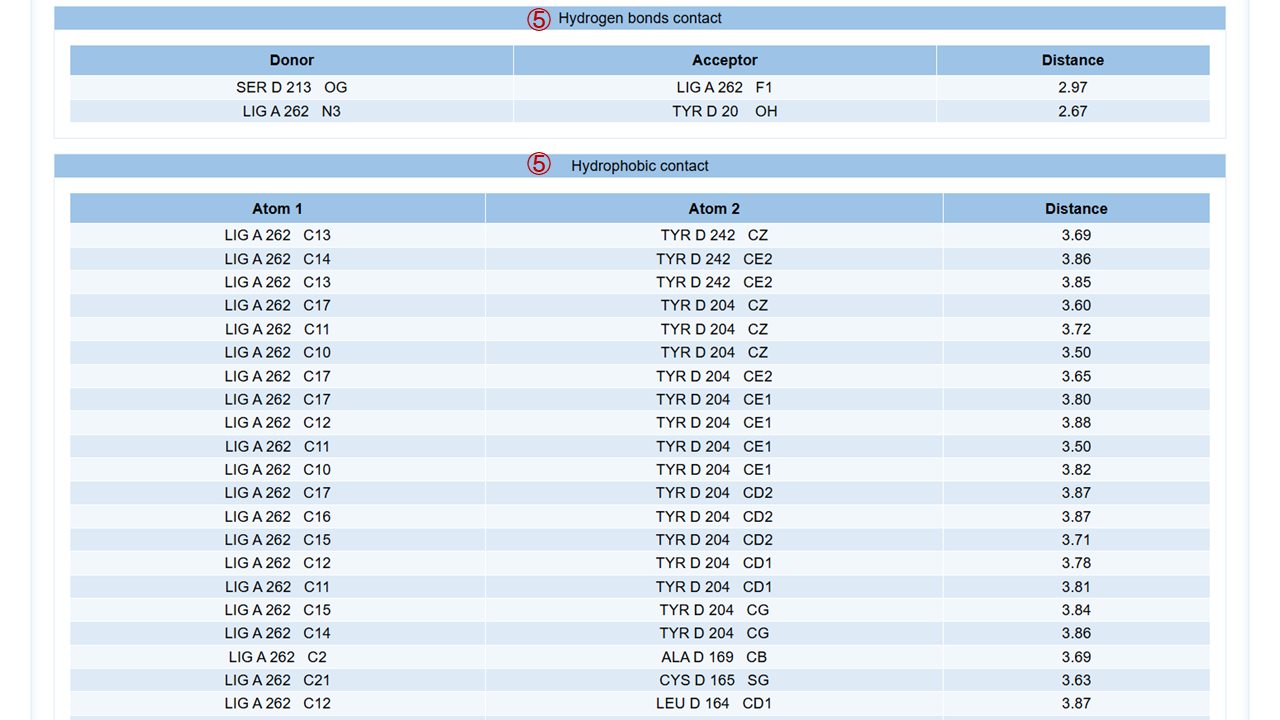
For each target, you can click to the detail button to view the detailed results, which include:(1) target general information; (2) protein physicochemical properties; (3) 2D binding mode diagram of the molecule and the target generated by Molscript, Pymol and Ligplot; (4) 3D binding mode diagram of the molecule and the target generated by Molstar, where you can freely rotate within the graphic frame and observe the interactions between the molecule and the target; (5) the hydrogen bonds and hydrophobic contact interaction. Here are some operation tips for Molstar:
Highlight: Hover element using mouse hover
Extend highlight: From selected to hovered element along polymer using mouse hover + shift key
Toggle extended selection: Click on element using left mouse button + shift key to extend selection along polymer
Toggle selection: Click on element using left mouse button
Deselect all: Click on nothing using left mouse button
Representation Focus: Click element using left mouse button
Representation Focus Add: Click element using left mouse button + shift key
Camera center and focus: Click element using left mouse button or right mouse button or left mouse button + control key
Camera center and focus: Click element using right mouse button or left mouse button + control key
Reset camera focus: Click on nothing using left mouse button or right mouse button or left mouse button + control key
Spin Animation: Press i
Rock Animation: Press o
Toggle Fly Mode: Press space + shift key
Reset View: Press t
Gobal Illumination: Press g
Standard SMILES format The SMILES format supported by this server is Daylight SMILES, you can find its details in here.